Le Technion (Israël) perce le mystère de l'évolution animale pour mieux comprendre le cancer
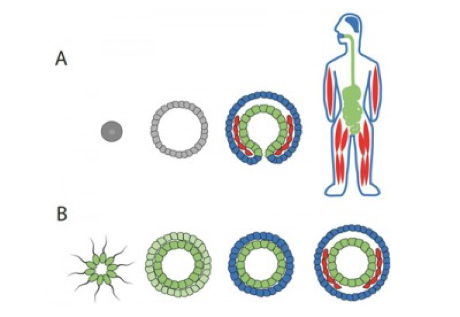
[:fr]La plupart des embryons d’origine animale contiennent trois couches de cellules qui se transforment en chaque partie du corps, des cerveaux, aux os et aux organes. Depuis le 19ème siècle, les biologistes se demandaient laquelle de ces couches est venue en premier dans l’évolution animale. Grâce à une puissante technologie développée au Technion, il est possible à présent déchiffrer les secrets de l’évolution, ce qui peut avoir un impact important pour aider à comprendre le mécanisme du cancer.
A lire sur technion.ac.il[:en]
Most animal embryos contain three layers of cells that transform into every part of the body—from brains to bones to guts. Since the 19th century, biologists have been puzzling over which of these layers came first in animal evolution. We now have the answer, thanks to a powerful 21st-century technology developed at the Technion and deciphering the secrets of evolution may have a side benefit in helping to understand cancer.
Using a method called CEL-Seq that can spy on the activity of every gene within a cell at once, Technion Associate Professor Itai Yanai and his colleagues now provide compelling evidence that the layer called the endoderm evolved first, followed by the ectoderm layer and finally the mesoderm layer.
Yanai has other big plans for CEL-Seq. “We applied this unbelievably powerful tool to figuring out the evolution of the germ layers, a 19th century problem, but it will also be useful in things like cutting-edge cancer research as well,” he said.
Yanai and his colleagues used CEL-Seq to show that genes turned on in the endoderm of a worm embryo are the first to turn on in development, followed by genes turning on in the ectoderm and then mesoderm. They also detected that endoderm genes are older than genes mostly found in the ectoderm and mesoderm.
In their paper published in the December 10 advance online edition of Nature, the researchers argue that the endoderm layer dates back to ancient single-celled organisms that banded together to form the first multicellular animals. Endoderm cells kept up their ancestral feeding function in the new animals, which freed up the other cells to evolve into new layers in the animal.
Understanding how evolution has altered cells in the past can also “reveal to us what is easily changeable and what is not changeable in a cell,” Yanai added. “If a cell goes into a disease state, for instance, we might know more about what we could do to reverse this state and what might be more difficult to reprogram in the cell.”
CEL-Seq was invented at Technion in 2012 by the Yanai lab. Yanai said that one way to envision the method’s power is to consider a single human cell and the 20,000 genes it contains as a room with 20,000 light switches.
“Each room can have a different mood, because we can turn on or turn off those switches in many different ways. This is why cells with the same genes can have many, many different types of behavior,” he explained. “With this method, we can take a given cell and know the position of every switch—whether it’s on or off–and from this infer what functions are going on.”
Currently a Fellow at the Radcliffe Institute of Advanced Study at Harvard University, Yanai is using the method to study the development of tumors in zebrafish. “For the last ten years I’ve been working on development and evolution, all seen through the eyes of gene expression,” he said, “and I’ve realized that cancer is actually a natural extension of this work since here you also have cells developing and evolving.”
Yanai’s team hasn’t finished unleashing CEL-Seq on other big questions in biology, either. One of their ongoing studies uses the method to look at all the genes in ten vastly different animal species and compare how they are regulated throughout the development of the embryo. “We want to see what makes an animal an animal, what is universal across all of them,” Yanai said.
[:]