Weizmann (Israël) : une analyse dynamique des populations bactériennes du corps humain

[:fr]
Comment mesurer le taux de croissance d’une population bactérienne ?
Ce taux de croissance est déterminé en analysant la réplication de l’ADN bactérien. En effet, une bactérie qui se divise doit au préalable dupliquer son ADN. Chez la plupart des bactéries, la réplication de l’ADN débute toujours au même endroit, appelé origine de réplication. Elle se poursuit de façon bidirectionnelle jusqu’à ce que les deux brins en croissance parviennent au « terminus » de réplication. Les portions de l’ADN qui ont déjà été répliquées (les plus proches de l’origine de réplication) seront donc présentes dans une bactérie en deux exemplaires, et le reste du génome en un exemplaire. Une fois que la réplication est terminée, la bactérie se divise en deux et chacune des deux bactéries ainsi formées hérite d’une copie du génome.
Les chercheurs ont procédé à un séquençage de nombreux échantillons bactériens en mesurant uniquement à chaque fois le nombre total de copies de l’origine de réplication et celui du terminus de réplication dans l’ensemble de la population. Ils analysent le ratio de ces deux nombres (nombre de copies de l’origine/nombre de copies du terminus), qu’ils appellent PTR (Peak to Trough, littéralement « pic sur creux »). Si une population bactérienne est stable, ce ratio doit être proche de 1, puisque chaque bactérie aura un exemplaire de l’origine de réplication et un exemplaire du terminus. Si la population se divise activement, ce ratio tendra vers deux, puisque la plupart des bactéries seront en train de répliquer leur génome et posséderont deux copies de l’origine de réplication, pour une copie du terminus. Les chercheurs ont montré qu’effectivement la mesure des PTR à un instant donné reflétait de façon très précise le taux de croissance des bactéries dans la demi-heure suivante.
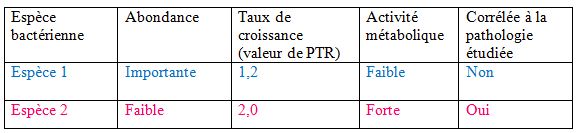
Quelles nouvelles informations la mesure du taux de croissance peut-elle apporter ?
Les chercheurs sont capables de mesurer le PTR spécifique d’une espèce donnée même quand elle se trouve au sein d’une communauté bactérienne. Ils ont ainsi pu étudier des échantillons humains et regarder s’il y avait des associations entre des pathologies et des variations de PTR –autrement dit des variations de taux de croissance- chez des espèces bactériennes spécifiques. Rechercher des corrélations entre la présence de certaines maladies et les espèces bactériennes présentes dans l’organisme n’est pas nouveau : on sait par exemple que certaines maladies infectieuses de l’intestin sont associées à un déséquilibre dans l’abondance relative de certaines espèces de bactéries dans le tube digestif. Cependant aucune étude dynamique n’avait été réalisée jusque là. Le présent travail a révélé des associations entre le taux de croissance de certaines espèces bactériennes et des pathologies comme le diabète de type II ou la maladie de Crohn. Les espèces qui possèdent un taux de croissance exceptionnellement élevé dans certaines circonstances pathologiques ne sont pas nécessairement les plus abondantes mais sont métaboliquement très actives (tableau ci-après).
Mis à part les connaissances nouvelles qu’elle a déjà commencé à apporter sur les dynamiques bactériennes en situation pathologique, l’utilisation de la mesure de PTR ouvre encore d’autres perspectives tout à fait enthousiasmantes. Notamment celle de pouvoir observer, en direct, quelles espèces réagissent rapidement à un traitement thérapeutique donné. Cette technique pourra aussi permettre de déterminer plus précisément les interactions entre espèces au sein du microbiome : quelles espèces réagissent en premier à certains stimuli (alimentaires, pathologiques, médicaux,…) et modulent la croissance des espèces voisines.
Auteur : Tirtsa Toledano pour BVST
Publication dans PNAS, 4 septembre 2015
[:en]
The group tested this formulation experimentally, first in single-strain cultures for which the growth rate could be controlled and observed, then in multiple animal model systems, and finally in the DNA sequences of human microbiomes, in their full complexity. Their method worked even better than expected: The estimated bacterial growth rates turned out to be nearly identical to observed growth rates. “Now we can finally say something about how the dynamics of our microbiome are associated with a propensity to disease. Microbial growth rate reveals things about our health that cannot be seen with any other analysis method,” says Elinav.
[:]